Student Attitudes Contribute to the Effectiveness of a Genomics CURE
We dedicate this paper to the memory of our colleague James Youngblom, who was a tireless advocate for students and was involved in all stages of creation of this work up until his untimely death in late June 2021.
Lopatto, David, et al. “Student Attitudes Contribute to the Effectiveness of a Genomics CURE.” Journal of Microbiology & Biology Education (2022): e00208-21.
Abstract
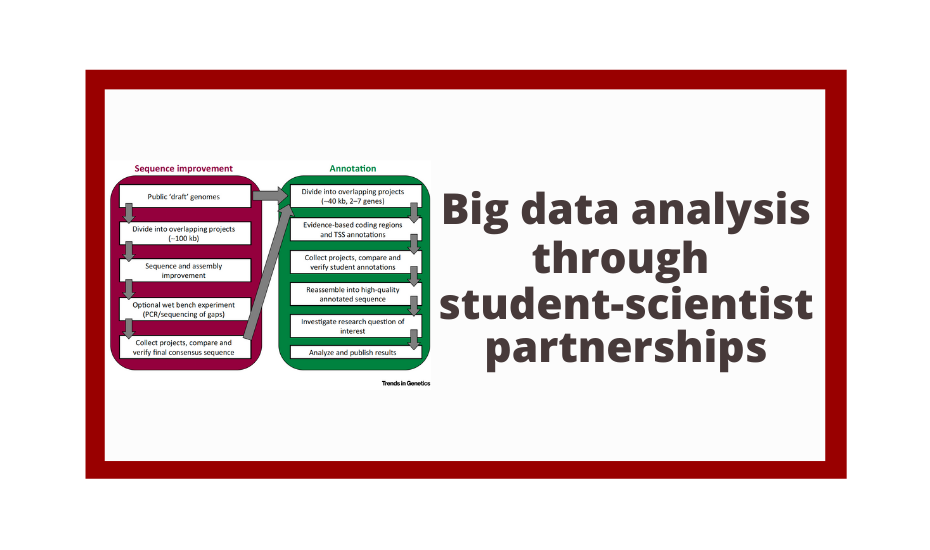
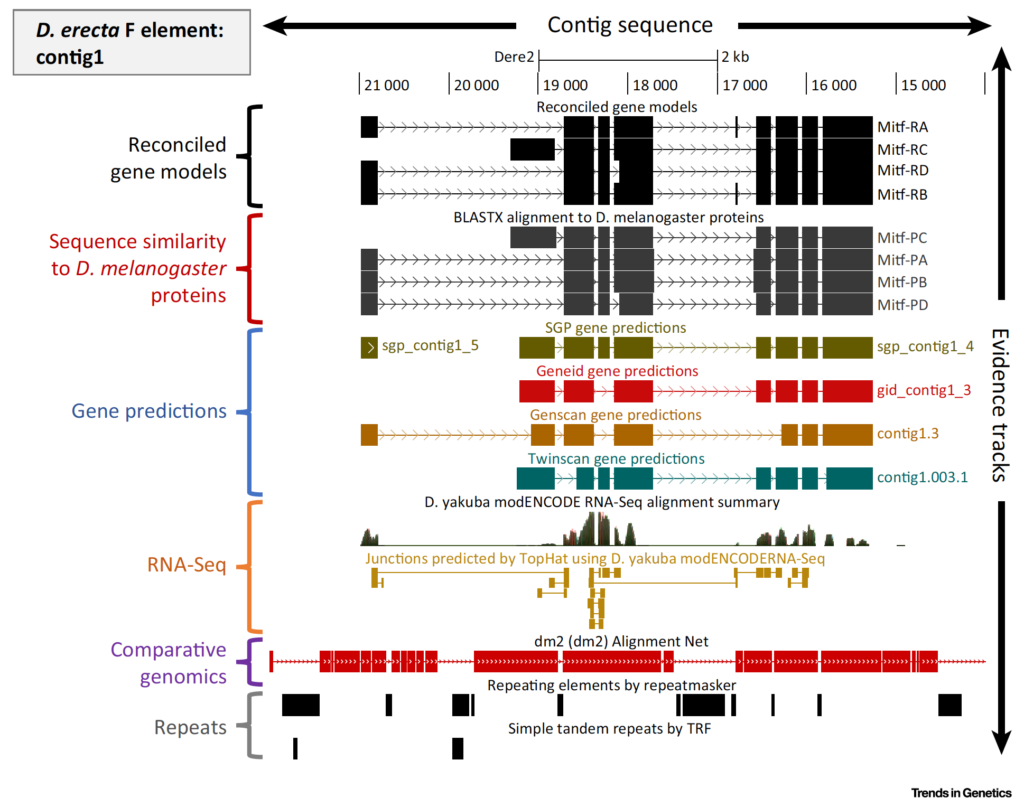
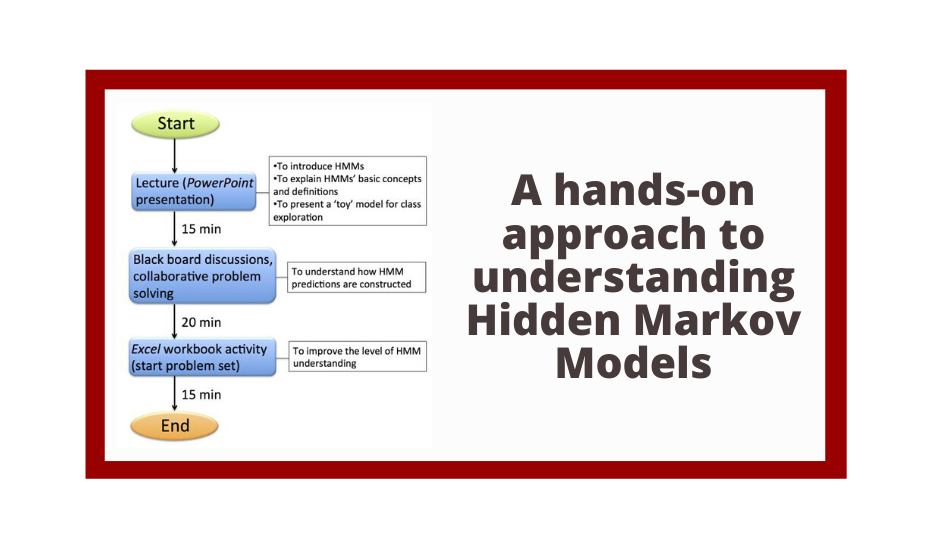
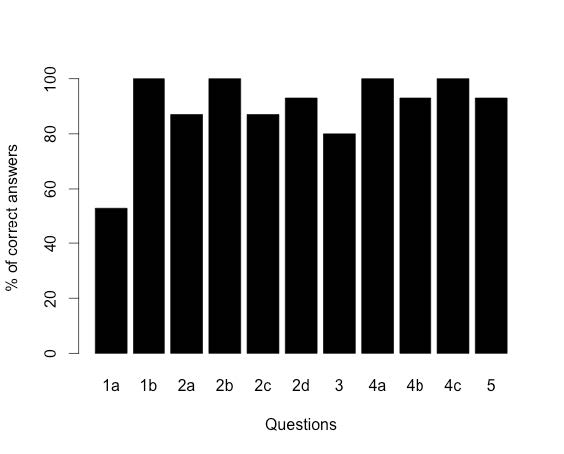
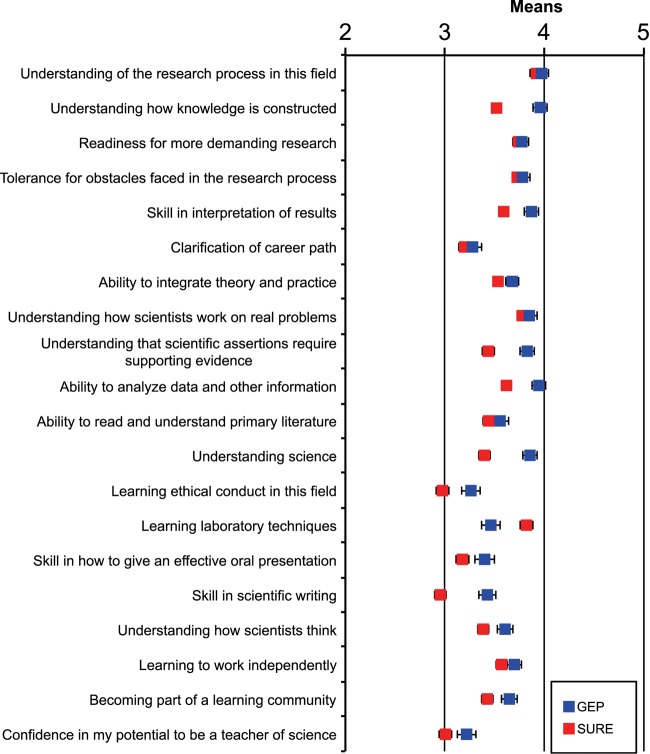
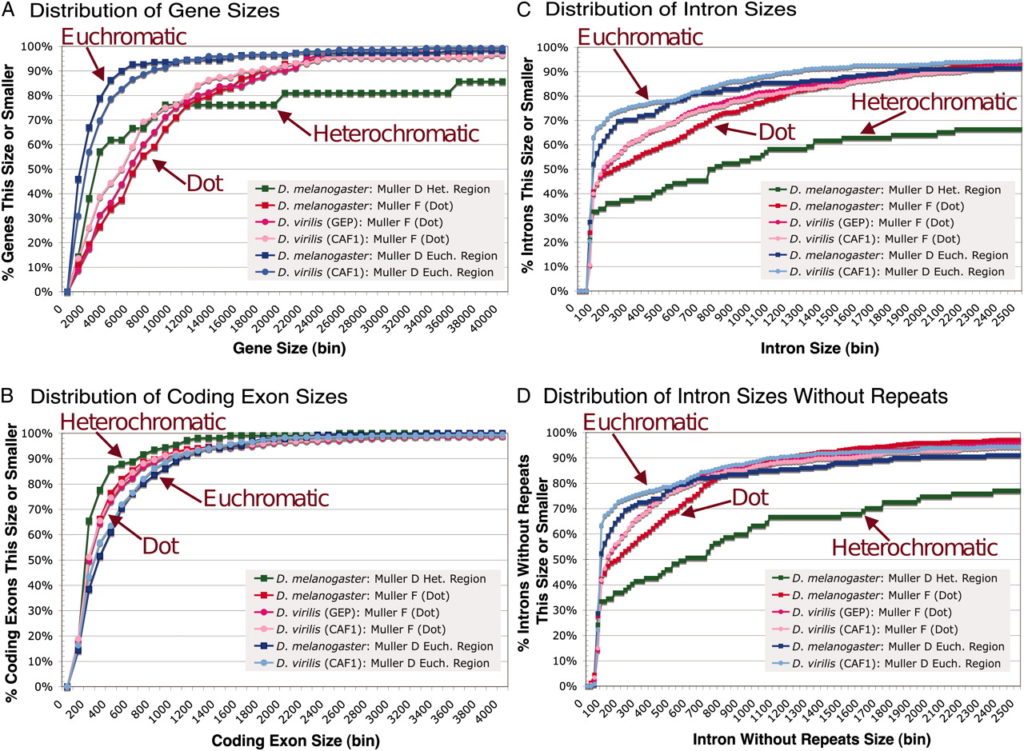
The Genomics Education Partnership (GEP) engages students in a course-based undergraduate research experience (CURE). To better understand the student attributes that support success in this CURE, we asked students about their attitudes using previously published scales that measure epistemic beliefs about work and science, interest in science, and grit. We found, in general, that the attitudes students bring with them into the classroom contribute to two outcome measures, namely, learning as assessed by a pre- and postquiz and perceived self-reported benefits. While the GEP CURE produces positive outcomes overall, the students with more positive attitudes toward science, particularly with respect to epistemic beliefs, showed greater gains. The findings indicate the importance of a student’s epistemic beliefs to achieving positive learning outcomes.