Carolinas Regional Node Meeting – April 22, 2022
The Carolinas Regional Node Meeting was held virtually on Friday, April 22, 2022, 9:00am-12:15pm (ET). The meeting kicked off with brief welcome remarks by Jeff French, a Node member from North Greenville University, SC, who also introduced our keynote speaker. Nate Mortimer from Illinois State University, who leads the Parasitoid Wasps Project in the GEP, gave an inspiring talk on “Invasion of the Body Snatchers: Parasitoid Wasps of Drosophila.” We had a break between 9:50-10am to allow for preparation for students’ presentations. Right before diving on these, Marisol Santisteban from UNC at Pembroke, current Carolinas Node leader, gave a brief introduction to the GEP and acquainted attendees with the organization: membership, different modes of participation, and the projects that are currently pursued.
Engaging presentations by five students from South Wesleyan University (Michelle Eller, advisor) and Appalachian State University (Clare Scott Chialvo, advisor) took us into the intricacies of their projects in the Insulin Signaling Pathways Project and in an emerging new project on detoxification genes, and the challenges of manual annotation of species closely related to D. melanogaster, such as D. immigrants. After a break from 11-11:15, the last hour of the day was a professional development event geared towards students. The “Careers in Genomics” panel hosted 5 experts from different Genomics fields, with different levels of education (not all PhDs), and not all from academia. All panelists were female and from diverse backgrounds. Four of the five panelists also work in North Carolina which shows our Carolinas students that there is a future for them in this field that’s also close to home. Sabrina Powell, Education Program Director of the Precision Medicine in Health Care in the Department of Genetics at UNC Chapel Hill School of Medicine generously helped assemble this extraordinary array of experts and prepared a fictional but realistic scenario about a 3-year-old boy diagnosed with autism, who is referred to the UNC Genetics Clinic for further testing. The testing reveals two specific variants in Mateo’s exome, one which is known to cause a specific subtype of autism and another which is associated with a high risk of adult-onset dementia. There was a role for professionals at each of our panelists’ positions:
- Kate Foreman, CGC, Genetic Counselor (University of North Carolina at Chapel Hill);
- Meghan Halley, PhD, MPH, Senior Research Scholar (Center for Biomedical Ethics, Stanford University);
- Julie Horvath, PhD, Head of Genomics & Microbiology Research Lab (NC Museum of Natural Sciences) and Research Associate Professor, Biological and Biomedical Sciences (NC Central University);
- Halina Krzystek, Bioinformaticist, Bioinformatics Data Services, Q-Squared Solutions; and
- Janae Simons, Bioinformatics Software Developer (Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill).
The meeting was attended by 34 people, of which 8 were undergraduate students, and three Community College instructors, two if which had expressed interest in knowing more about GEP and had been invited to New Member Trainings.
What worked well for your event that might help others plan similar events?
Using the opportunity that came with the “online” format we were able to host an outstanding keynote speaker (our very own Nate Mortimer from Illinois State University) and panel of experts in different fields of genomics for our “Careers in Genomics” panel; many were in North Carolina, but we had a person from Stanford. It would not have been possible to assemble this array of experts if it had been an in-person event. I would recommend some “professional development” event for students, like the “Careers in Genomics” panel that we had. We hosted individuals in different fields, with different education degrees (not all PhDs), and not all from academia. I believe the students really found this session interesting and useful. Making a program and emailing it to all participants was a plus and we also emailed certificates to the student presenters which is a nice touch of appreciation.
What lessons were learned from challenges in the planning or execution of the event that might be helpful to others?
It is hard to come up with the “perfect” date. We considered weekday vs. weekend, all day vs. half a day, mid semester vs. late in the semester. We finally settled on a Friday because they tend to be lighter days for students, and only half a day (morning). We did it towards the end of the semester, so students would have made enough progress in their projects to present. Considering that Node meetings will be in-person in the future, I would recommend a weekend, maybe start on Friday evening with posters and maybe the keynote speaker and then talks on Saturday morning and some professional development event. Some people may choose to attend only one day but they won’t be a full day. Or maybe make it a whole day event on Saturday. Keeping some form of hybrid might be useful, especially for the keynote speaker or panels, because that allows inviting speakers that are not geographically close to the Node.
As far as the execution, it is still hard to keep everyone engaged and have them turn on their cameras and ask questions. So as much as possible, I would recommend holding the student presentations in-person and if online, encourage folks to have their cameras on, and incentivize asking question with some form of reward. We provided a short bio of the panelist in the program that was emailed to the registrants the night before. If possible, do that earlier, so people may think of some questions to ask ahead, and openly ask them to try to do so.
Some students reported not receiving a link for the meeting, but they registered only minutes before the start of the meeting. If you plan to leave the registration open till the very start of the meeting, make sure someone in the Node does a last sweep. Ask the GEP staff to give access to registration to someone in the Node.
Keep the GEP staff in the loop for everything planning, they are incredible resourceful, helpful and efficient!
Southeast Regional Node Meeting April 22, 2022
Twelve undergraduate students shared their recent research annotating genes as part of the Pathways Project at the third semi-annual virtual Southeast Regional Node Meeting and research symposium on April 22, 2022. The event began with an engaging talk by Dr. Rebecca Varney, Postdoc at the University of California—Santa Barbara, on how gene annotation efforts can be especially helpful when working with non-model organisms.

Drosophila yakuba – Tsc1
Lose, B; Myers, A; Fondse, S; Alberts, I; Stamm, J; Youngblom, JJ; Rele, CP; Reed, LK (2021). Drosophila yakuba – Tsc1. microPublication Biology. 10.17912/micropub.biology.000474.
GEP Program Director Wins Outstanding Professor Award
We are pleased to announce that GEP’s Program Director—Dr. Laura Reed—was awarded the Outstanding Professor Award at The University of Alabama.
Read the press release for more information.
2021 Pathways GEP Summer Research Fellows
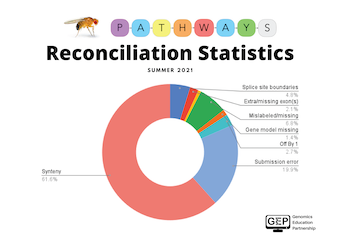
Collectively, the Summer Fellows reconciled 102 gene models. Approximately 10% of the submitted gene models were in congruence with the final gene models. The most common annotation error in the submitted gene models was improperly assigning synteny (62%). The image above illustrates the preliminary reconciliation statistics for the types of annotation errors for the Pathways Project in Summer 2021.
Reconcilers primarily focused on a single gene each, so that they could perform deeper analysis on the evolution of that gene within our selected clade. Presentations summarizing each of their work are publicly available on Box.
The Summer Fellows also generated microPublication documents. Multiple models of four genes (Tor, Ilp3, raptor, and Dsor1) will be sent to microPublication for review by the end of September 2021. Five of the reconcilers will continue to work with the Pathways Project team to reconcile gene models and send them for publication.
GEP Student Wins 1st Place at Research Conference
The University of Alabama’s 14th annual Undergraduate Research and Creative Activity Conference (URCA) was held on March 31, 2021.
Six GEP students (mentored by Dr. Laura Reed) presented at URCA 2021 one of which, Samantha Hoffman, won 1st place in her division.
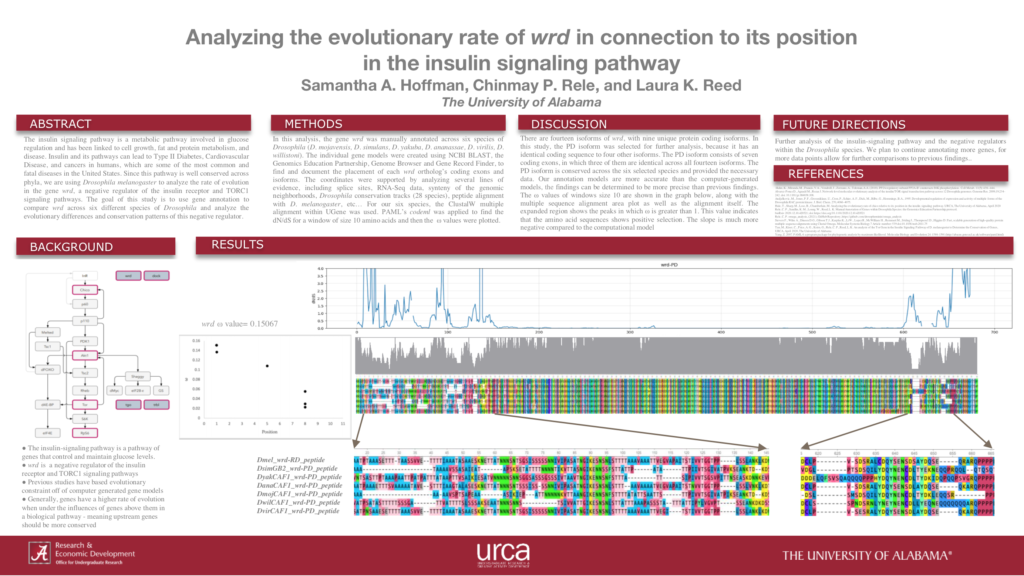
Analyzing the evolutionary rate of wrd in connection to its position in the insulin signaling pathway
Abstract: The insulin signaling pathway is a metabolic pathway involved in glucose regulation and has been linked to cell growth, fat and protein metabolism, and disease. Insulin and its pathways can lead to Type II Diabetes, Cardiovascular Disease, and cancers in humans, which are some of the most common and fatal diseases in the United States. Since this pathway is well conserved across phyla, we are using Drosophila melanogaster to analyze the rate of evolution in the gene wrd, a negative regulator of the insulin receptor and TORC1 signaling pathways. The goal of this study is to use gene annotation to compare wrd across six different species of Drosophila and analyze the evolutionary differences and conservation patterns of this negative regulator.
GEP’s Prototype microPublication is Published
GEP’s first/prototype microPublication has been published! Now that we have made it all the way through the process with the first one we will be moving quickly to submit the additional manuscripts that are ready to go.
Rele, CP; Williams, J; Reed, LK; Youngblom, JJ; Leung, W (2021). Drosophila grimshawi – Rheb. microPublication Biology. 10.17912/micropub.biology.000371.
GEP Annotation Protocol
Preprint
We’re excited to announce this preprint as it moves us one step closer to rolling out microPublications for the Pathways Project!
Abstract: Annotating the genomes of multiple organisms allows us to study their genes as well as the evolution of those genes. While many eukaryotic genome assemblies already include computational gene predictions, these predictions can benefit from review and refinement through manual gene annotation. The Genomics Education Partnership (GEP; thegep.org) has developed an annotation protocol for protein-coding genes that enables undergraduate students and other researchers to create high-quality gene annotations that can be utilized in subsequent scientific investigations. For example, this protocol has been utilized by the GEP faculty to engage undergraduate students in the comparative annotation of genes involved in the insulin signaling pathway in 28 Drosophila species, using D. melanogaster as the informant genome. Students construct gene models using multiple lines of computational and experimental evidence including expression data (e.g., RNA-Seq), sequence similarity (e.g., BLAST, multiple sequence alignments), and computational gene predictions. For quality control, each gene is annotated by at least two students working independently, followed by reconciliation of the submitted gene models by a more experienced student. This article provides an overview of the annotation protocol and describes how discrepancies in student submitted gene models are resolved to produce a final, high-quality gene set suitable for subsequent analyses. This annotation protocol can be adapted to other scientific questions (e.g., expansion of the Drosophila Muller F element) and other species (e.g., parasitoid wasps) to provide additional opportunities for undergraduate students to participate in genomics research. These student annotation efforts can substantially improve the quality of gene annotations in publicly available genomic databases.
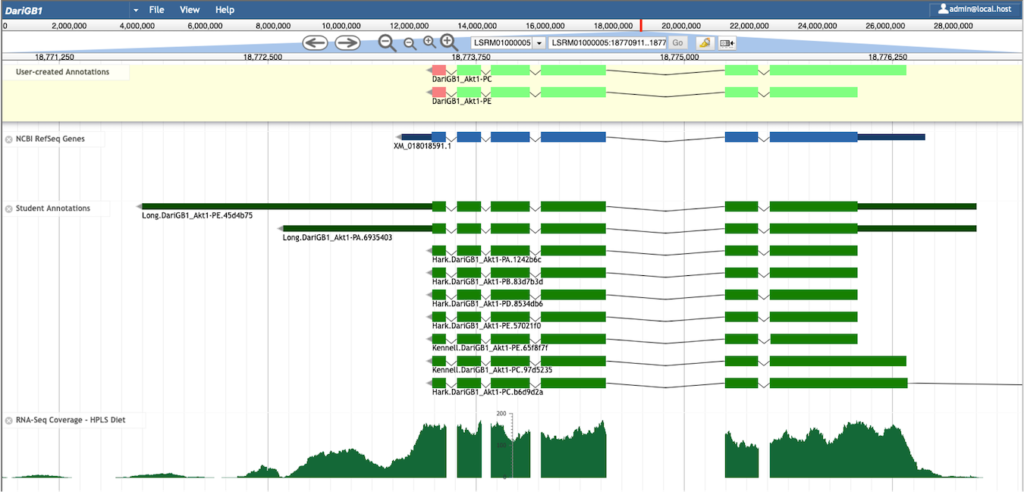
Rele CP, Sandlin KM, Leung W, Reed LK. Manual Annotation of Genes within Drosophila Species: the Genomics Education Partnership protocol. bioRxiv 2020.12.10.420521; doi: https://doi.org/10.1101/2020.12.10.420521.
Pathways Project: Annotation Workflow
The Annotation Workflow is a one page summary of the annotation protocol for the Pathways Project. This workflow provides an overview of the key analysis steps and bioinformatics tools for the annotation of a putative ortholog.
- « Previous
- 1
- 2
- 3
- Next »