We’re excited to announce this preprint as it moves us one step closer to rolling out microPublications for the Pathways Project!
Abstract: Annotating the genomes of multiple organisms allows us to study their genes as well as the evolution of those genes. While many eukaryotic genome assemblies already include computational gene predictions, these predictions can benefit from review and refinement through manual gene annotation. The Genomics Education Partnership (GEP; thegep.org) has developed an annotation protocol for protein-coding genes that enables undergraduate students and other researchers to create high-quality gene annotations that can be utilized in subsequent scientific investigations. For example, this protocol has been utilized by the GEP faculty to engage undergraduate students in the comparative annotation of genes involved in the insulin signaling pathway in 28 Drosophila species, using D. melanogaster as the informant genome. Students construct gene models using multiple lines of computational and experimental evidence including expression data (e.g., RNA-Seq), sequence similarity (e.g., BLAST, multiple sequence alignments), and computational gene predictions. For quality control, each gene is annotated by at least two students working independently, followed by reconciliation of the submitted gene models by a more experienced student. This article provides an overview of the annotation protocol and describes how discrepancies in student submitted gene models are resolved to produce a final, high-quality gene set suitable for subsequent analyses. This annotation protocol can be adapted to other scientific questions (e.g., expansion of the Drosophila Muller F element) and other species (e.g., parasitoid wasps) to provide additional opportunities for undergraduate students to participate in genomics research. These student annotation efforts can substantially improve the quality of gene annotations in publicly available genomic databases.
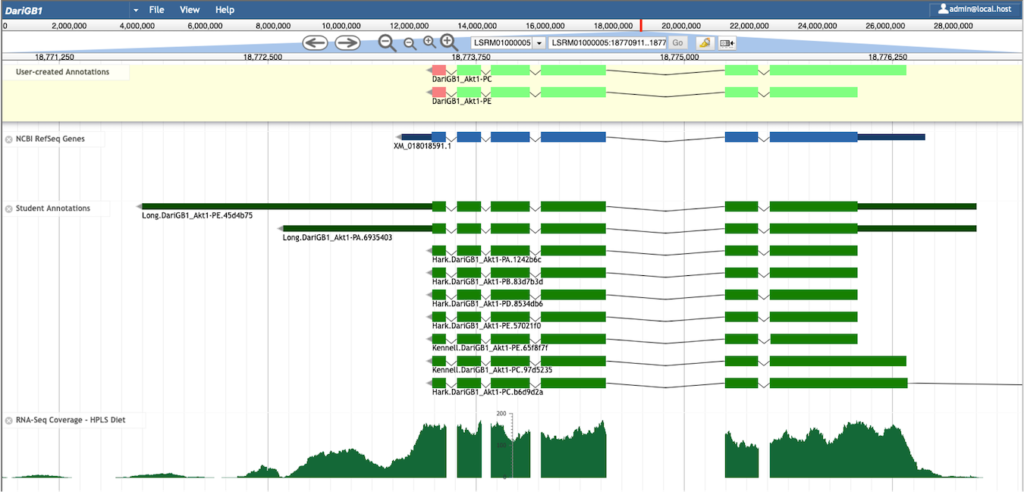
Rele CP, Sandlin KM, Leung W, Reed LK. Manual Annotation of Genes within Drosophila Species: the Genomics Education Partnership protocol. bioRxiv 2020.12.10.420521; doi: https://doi.org/10.1101/2020.12.10.420521.

