During Summer 2021, nine Summer Fellows from The University of Alabama and Oklahoma Christian University reconciled coding region annotation projects from the Pathways Project. The Summer Fellows—James O’Brien, Ryan Dufur, Aidan Long, Cole A. Kiser, Ashley Morgan, Alyssa Koehler, Annie Backlund, Jhilam Dasgupta, and Rachael Cowan—were mentored by Dr. Laura K. Reed with support from Chinmay P. Rele both of which are at The University of Alabama.
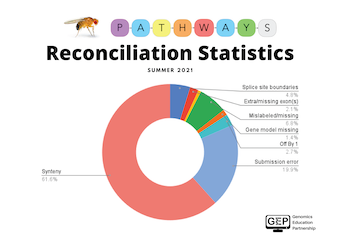
Collectively, the Summer Fellows reconciled 102 gene models. Approximately 10% of the submitted gene models were in congruence with the final gene models. The most common annotation error in the submitted gene models was improperly assigning synteny (62%). The image above illustrates the preliminary reconciliation statistics for the types of annotation errors for the Pathways Project in Summer 2021.
Reconcilers primarily focused on a single gene each, so that they could perform deeper analysis on the evolution of that gene within our selected clade. Presentations summarizing each of their work are publicly available on Box.
The Summer Fellows also generated microPublication documents. Multiple models of four genes (Tor, Ilp3, raptor, and Dsor1) will be sent to microPublication for review by the end of September 2021. Five of the reconcilers will continue to work with the Pathways Project team to reconcile gene models and send them for publication.
Collectively, the Summer Fellows reconciled 102 gene models. Approximately 10% of the submitted gene models were in congruence with the final gene models. The most common annotation error in the submitted gene models was improperly assigning synteny (62%). The image above illustrates the preliminary reconciliation statistics for the types of annotation errors for the Pathways Project in Summer 2021.
Reconcilers primarily focused on a single gene each, so that they could perform deeper analysis on the evolution of that gene within our selected clade. Presentations summarizing each of their work are publicly available on Box.
The Summer Fellows also generated microPublication documents. Multiple models of four genes (Tor, Ilp3, raptor, and Dsor1) will be sent to microPublication for review by the end of September 2021. Five of the reconcilers will continue to work with the Pathways Project team to reconcile gene models and send them for publication.