An Introduction to NCBI BLAST
This walkthrough serves as an introduction to key functionalities of NCBI BLAST. Exercise Exercise Worksheet Worksheet Answer Key Answer Key Package without Answers Package
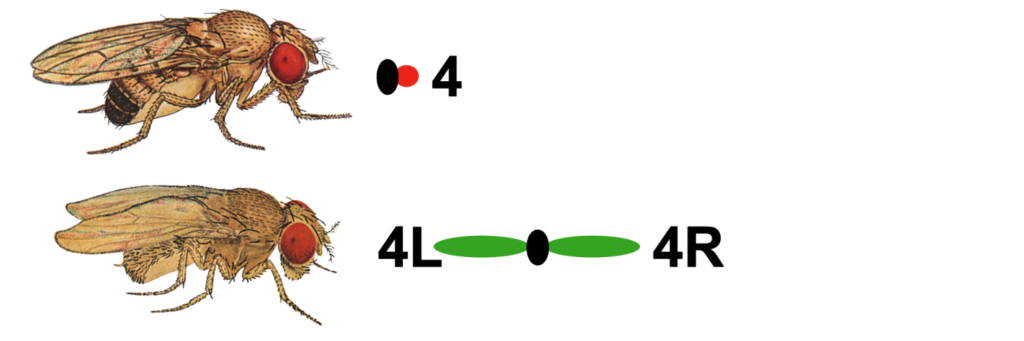
One of the unusual features of eukaryotic genomes is the discordance between genome size and the complexity of the organism (i.e., the C-value paradox; Eddy, 2012). The smallest chromosome in Drosophila melanogaster is chromosome 4 (also known as the Muller F element), with an estimated size of ~5.2 Mb (Locke and McDermid, 1993). The D. melanogaster F element is generally packaged as heterochromatin: it has a high repeat content, is packaged throughout with HP1a and H3K9me2/3; it exhibits late replication and little or no recombination. However, the banded portion (~1.4 Mb) of this chromosome also contains ~80 protein-coding genes. These F element genes exhibit a range of expression levels similar to genes that reside in euchromatic domains — indicating that F element genes have acquired distinct features that enable them to function in a heterochromatic environment (reviewed in Riddle and Elgin, 2018).
While the F element has maintained a similar size in many other Drosophila species, it is substantially larger in at least four Drosophila species (i.e., D. ananassae, D. bipectinata, D. kikkawai, and D. takahashii). For example, the D. ananassae Muller F element is more than 18.7 Mb in size. This study will examine the factors (e.g., transposon density) that have contributed to the expansion of the F element in these four Drosophila species, and assess the impact of this expansion on gene characteristics (e.g., codon bias, intron size).
GEP students will produce coding region and transcription start site annotations for F element genes in D. ananassae, D. bipectinata, D. kikkawai, and D. takahashii, as well as for genes in a euchromatic reference region derived from the Muller D element. (Euchromatic regions have not expanded in these species.) Comparative analyses using these datasets will provide insights into the evolutionary impacts of changes in chromosome and gene size, and will facilitate the identification of factors that enable genes to function in a heterochromatic environment. We anticipate that this work will move us toward a better understanding of how and why eukaryotic genomes became so large, for mammals, ~1000X larger than that of E. coli.
This walkthrough serves as an introduction to key functionalities of NCBI BLAST. Exercise Exercise Worksheet Worksheet Answer Key Answer Key Package without Answers Package
An introductory exercise using BLAST to annotate a region in the Drosophila melanogaster genome. Students can use this exercise to gain familiarity with performing BLAST searches and interpreting BLAST output. An answer key is provided for instructors.
Dr. Justin R. DiAngelo (Penn State Berks) and Dr. Alexis Nagengast (Widener University) have developed an exercise that introduces students to the basic functionality of the NCBI web site and NCBI BLAST. Students will use NCBI BLAST to identify the putative orthologs of the human Leptin gene in other species.
Developed by Jeremy Buhler, this PowerPoint presentation provides an introduction to the core algorithms that form the basis for efficient mapping of RNA-Seq reads against a genome or transcriptome. The video that accompanies this presentation was developed by Leocadia Paliulis (Bucknell University). PowerPoint Handout
This exercise continues your introduction to practical issues in comparative annotation. You will be annotating genomic sequence from the dot chromosome of Drosophila mojavensis using your knowledge of BLAST and some improved visualization tools. You will also consider how best to integrate information from high-throughput sequencing of expressed RNA.
This PowerPoint presentation provides a brief primer on the recommended annotation strategy for Drosophila projects. The presentation provides an overview of the goals of the GEP annotation project, an introduction to RNA-Seq, web databases, and a discussion on the phases of the splice donor and acceptor sites.
This walkthrough uses the annotation of a gene on the D. biarmipes Muller F element to illustrate the GEP comparative annotation strategy. This document shows how you can investigate a feature in an annotation project using FlyBase, the Gene Record Finder, and the gene prediction and RNA-Seq evidence tracks on the GEP UCSC Genome Browser. The walkthrough then shows how you can identify the coordinates of each coding exon using NCBI BLAST, and also includes a discussion on the phases of the donor and acceptor splice sites. The walkthrough concludes by verifying the proposed gene model using the Gene Model Checker; it also includes a sample GEP Annotation Report.
This worksheet will guide you through a series of basic steps that have been found to work well for annotation of species closely related to Drosophila melanogaster. It provides a technique that can also be the foundation of annotation in other, more divergent species.
This document is the revised annotation report that GEP students will use to report their annotation results to the GEP.
This workflow provides an overview of the key analysis steps and bioinformatics tools for the annotation of a predicted gene in the Drosophila F element GEP project.
A one-page summary/flowchart of the logic process for identifying appropriate splice sites when annotating.
This decision tree illustrates the list of criteria that can be used to determine the putative D. melanogaster ortholog of a predicted gene.
Similar to the Lecture Notes on Alignment, this is a PowerPoint presentation given by Dr. Jeremy Buhler for the GEP faculty and TA workshops. This presentation covers the basics of alignment, essential for students to correctly interpret BLAST results.
Notes from a lecture on sequence alignment given by Dr. Jeremy Buhler in the Bio 4342 course at WU. The lecture covers the theory behind BLAST as well as some of the potential problems and limitations of BLAST.
This PowerPoint presentation describes the recommended annotation strategy for Drosophila projects. The presentation provides an overview of the goals of the GEP annotation project, an introduction to NCBI BLAST, web databases, and the issue of reading frames and phase.
This is a PowerPoint presentation describing the recommended strategies for annotating a D. virilis fosmid. The homology-based annotation strategy should also be applicable to annotation of D. erecta and D. mojavensis projects.
This document is a more in depth description of the evidence based annotation technique used by the GEP. This document is designed to complement and extend the basic technique described in the Annotation for D. virilis PowerPoint.
This document illustrates how the strategies outlined in the Annotation Instruction Sheet can be applied to more challenging annotation cases.
Developed by Dr. Nick Reeves at Mt. San Jacinto College, Menifee Valley Campus, this PowerPoint presentation provides a brief overview of the Digital Lab Notebook, which provides detailed guidance to students on the GEP annotation strategy.
This presentation illustrates the unusual genomic features that GEP students have encountered as part of their annotation of Muller F Elements from Drosophila ananassae and D. bipectinata. The Muller F Elements in these two species have undergone substantial expansion compared to D. melanogaster. The presentation describes the basic strategy for identifying pseudogenes, retrogenes, partial gene duplications, pseudogene clusters, and nuclear mitochondrial DNA segments (NUMT) within these F Element annotation projects.
This PowerPoint presentation describes the common errors observed in student annotations.
Developed by Dr. Jeremy Buhler, this PowerPoint presentation provides an overview of the approaches for quantifying transcript abundance based on RNA-Seq data. The presentation includes a discussion on the benefits and limitations of the two approaches commonly used for RNA quantitation – RPKM and TPM.
This document describes the primary annotation goals to be included in the final oral presentation and written report for students enrolled in the Bio4342 course at WU.
This PowerPoint presentation describes the recommended annotation strategy for identifying transcription start sites in Drosophila. The presentation provides an overview of the promoter architecture in D. melanogaster and describes the types of evidence that can be used to support the transcription start sites annotations.
This walkthrough illustrates the GEP protocol for the comparative annotation of transcription start sites (TSS) in D. biarmipes. The walkthrough also includes a sample GEP TSS Report for the TSS annotation of onecut.
This workflow provides an overview of the key steps and recommended search parameters for the annotation of transcription start sites.
This document contains the notes from a lecture on motif finding given by Dr. Jeremy Buhler in the Bio 4342 course at WU. The lecture covers the different approaches used to represent sequence motifs and to search for sequence motifs in a genome.
Developed by Dr. Jeremy Buhler, this exercise uses MEME to discover putative regulatory motifs in a collection of D. melanogaster promoter sequences. It also illustrates some of the challenges associated with motif finding and the limitations of motif finding programs.
This walkthrough uses FlyBase, FlyFactorSurvey, and Patser to identify transcription factor binding sites in the region surrounding the transcription start site of onecut in D. biarmipes.
This walkthrough uses FlyBase RNA-Seq Search and the MEME suite to discover motifs that are enriched in a collection of D. melanogaster Muller F element genes that show similar expression patterns.
This lecture introduces students to the analysis of repetitious elements in the genome. It can be used as a stand-alone lecture, or included in the “F Element Project: Annotated Lecture Slides” sequence of lectures.
Similar to the lecture notes on Repetitious DNA, this is a PowerPoint presentation given by Dr. Jeremy Buhler for the GEP faculty and TA workshops. This presentation covers the basics of RepeatMasker, as well as limitations of the program that students should be aware of.