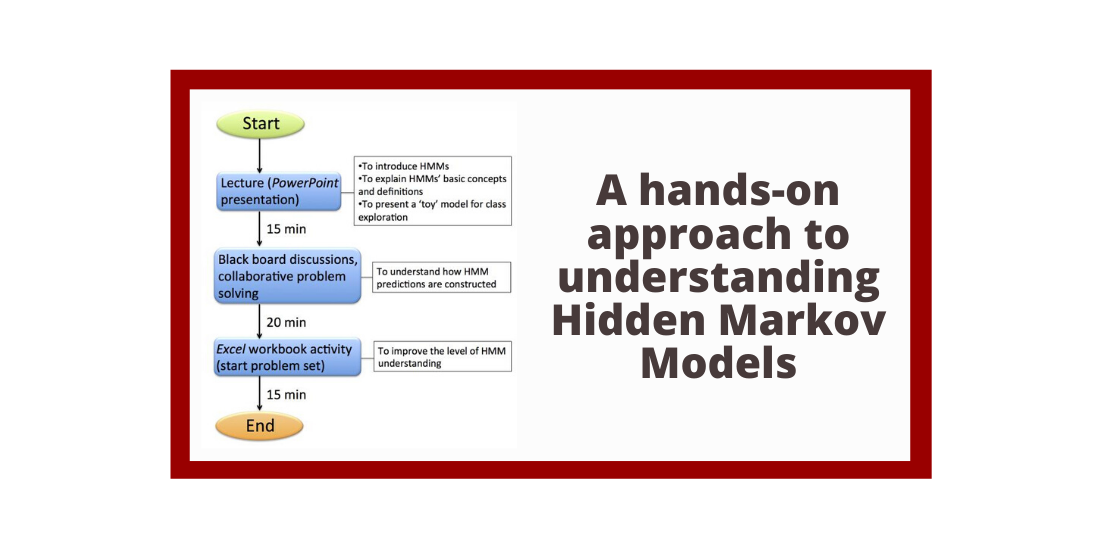
Abstract: In this Lesson, we describe a classroom activity that demonstrates how a Hidden Markov Model (HMM) is applied to predict a eukaryotic gene, focusing on predicting one exon-intron boundary. This HMM lesson is part of the BIOL/CS 370 ‘Introduction to Bioinformatics’ course (Truman State University, MO) and of Bio4342 ‘Research Explorations in Genomics’ (Washington University in St. Louis, MO). The original target student audiences include both Biology and Computer Sciences majors in their junior and senior years, although we believe the model activity would be successful with younger students. The class session starts with a brief introductory lecture describing HMMs and the terminology used in defining the parameters for a given model. This lecture is followed by students’ exploration of the HMM using Excel spreadsheets to manage calculations while they alter the key variables; collaborative problem solving and discussion of their strategies and results; and homework to check their understandings.
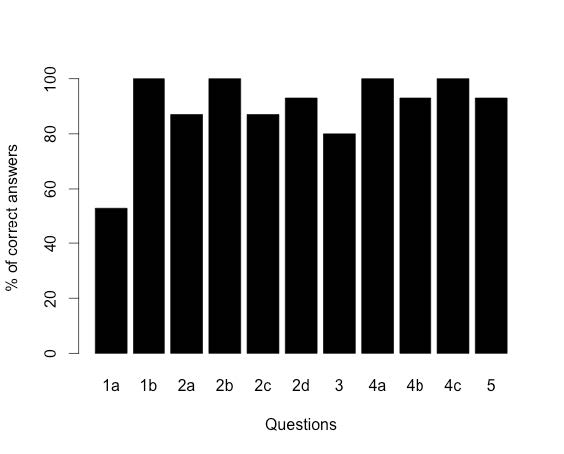
Students have reacted very positively to the HMM curriculum. Students with more computer science experience tended to ask more questions concerning the model itself. Overall, students performed well on the homework assignment, leading us to believe that we are a step closer to our main goal of filling the intellectual gap between computer scientists and biologists.
Weisstein, A.E., Gracheva, E., Goodwin, Z., Qi, Z., Leung, W., Shaffer, C.D. and Elgin, S.C.R. 2016. A Hands-on Introduction to Hidden Markov Models. CourseSource. https://doi.org/10.24918/cs.2016.8