Introduction to Pathways Project
Lecture is designed to introduce students to the big picture of the Pathways Project.
Biological systems are networks, and in these networks, we can define nodes (e.g., genes, proteins, metabolites) connected through edges (e.g., enzymatic/chemical reactions, transcription regulation). Networks have properties that can be measured using a mathematical approach, and we can make predictions about the evolution of a system based on some of those properties.
A “pathway” in a biological system can be defined as a relatively discrete (though never completely isolated) portion of a network. Generally, we view a pathway as a sequence of gene regulatory and enzymatic reactions that produce some important biological outcomes (e.g., synthesize an energy storage molecule, sense and regulate blood sugar levels).
In this project we will be using network analysis approaches to better understand the evolution and function of biological pathways. The Pathways Project is focused on annotating genes found in well characterized signaling and metabolic pathways across the Drosophila genus. The current focus is on the insulin signaling pathway which is well conserved across animals and critical to growth and metabolic homeostasis. The long-term goal of the Pathways Project is to analyze how the regulatory regions of genes evolve in the context of their positions within a network and we anticipate that other pathways will eventually be part of the analyses.
Pathways Project Overview provided by the Project Leader, Laura K. Reed (6 minutes) Slideset
The Pathways Project uses network analysis approaches to better understand the evolution and function of biological pathways. This GEP project is focused on annotating genes found in well characterized signaling and metabolic pathways across the Drosophila genus. The current focus is on the insulin signaling pathway which is well conserved across animals and critical to growth and metabolic homeostasis. The long-term goal of the Pathways Project is to analyze how the regulatory regions of genes evolve in the context of their positions within a network and we anticipate that other pathways will eventually be part of the analyses.
Lecture is designed to introduce students to the big picture of the Pathways Project.
This walkthrough illustrates how to apply the GEP annotation strategy for the Pathways Project to construct a gene model for the Ras homolog enriched in brain (Rheb) gene in Drosophila yakuba.
The Annotation Workflow is a one page summary of the annotation protocol for the Pathways Project.
The Reference Glossary includes definitions for terms that are frequently used in the Pathways Project.
This “Annotation Form” merged the “Annotation Report” and “Annotation Notebook” into a single document and the latter two items are now archived.
The Annotation Form Exemplar is provided as an example of a completed Annotation Form ready for submission to the GEP’s Pathways Project. The optional questions were omitted from the exemplar.
Students can apply what they learned in the Annotation Walkthrough to construct a gene model for Rheb in D. pseudoobscura by completing the Pathways Project: Annotation Form. This answer key is provided to assist instructors in checking the accuracy of the annotation and includes potential areas of confusion throughout.
This series of videos is intended to help GEP students annotate a Pathways Project gene from start to finish.
This was created in response to a member mentioning their students really struggled with the genomic neighborhood and the member didn’t realize until they were already too far into the annotation to correct their misconceptions. This is meant to be a quick in-class and/or homework assignment.
This resource is a slide deck offering an expanded introduction to synteny. Instructors are encouraged to use and modify to fit the needs of specific courses and students.
This PowerPoint presentation provides a primer on the recommended annotation strategy for the Pathways Project. The presentation provides an overview of the goals of the Pathways Project annotations, an introduction to RNA-Seq, web databases, and a discussion on the phases of the splice donor and acceptor sites.
This lesson and exercise defines similarity in a non-biological and biological sense, quantifies the similarity between two sequences, explains how a substitution matrix is used to quantify similarity, calculates amino acid similarity scores using the BLOSUM 62 substitution matrix, explains how BLAST detects similarity between two sequences and how to use
This module introduces students to the GEP UCSC Genome Browser. After completing this module students will be able to navigate to a genomic region and to control the display setting for different evidence tracks.
This module uses mRNA data to identify splice sites. After completing this module students will be able to identify intron-exon boundaries using canonical splice donor and acceptor sequences and determine which are best supported by RNA-Seq and TopHat splice junction predictions.
In this module students will learn how mRNA is translated into a string of amino acids. After completing this module students will be able to determine the codons for specific amino acids as well as start and stop codons. They will be able to identify open reading frames for a given gene, define the phases of splice donor and acceptor sites and describe how they impact the maintenance of the open reading frame.
This module explores how multiple different mRNAs and polypeptides can be encoded by the same gene. After completing this module students will be able to explain how alternative splicing of a gene can lead to different mRNAs and illustrate how alternative splicing can lead to the production of different polypeptides and result in drastic changes in phenotype.
This walkthrough serves as an introduction to key functionalities of NCBI BLAST. Exercise Exercise Worksheet Worksheet Answer Key Answer Key Package without Answers Package
This PowerPoint presentation provides a brief introduction to the different types of RNA-Seq evidence tracks (e.g. Bowtie, TopHat, Cufflinks) that are on the GEP UCSC Genome Browser.
In this example, Ilp6 is within the intron of Raf-PE, however Raf-PA is upstream of Ilp6.
We are defining gene order based on the first/closest protein coding exon only. So if the gene is nested in an intron that is between two non-coding exons then we ignore those UTRs and just define gene order based on the coding exons. If a gene is nested in an intron between two coding exons of another gene then we describe that as nesting. So in this example, Raf is upstream of Ilp6
The Genome Browser Gateway should default to the correct assembly once you click on the Drosophila species in the left-hand table. To double check, you are using the correct one, you can see which assembly you should be using via the “Genome Browsers” column of the Pathways Project Genome Assemblies web page. 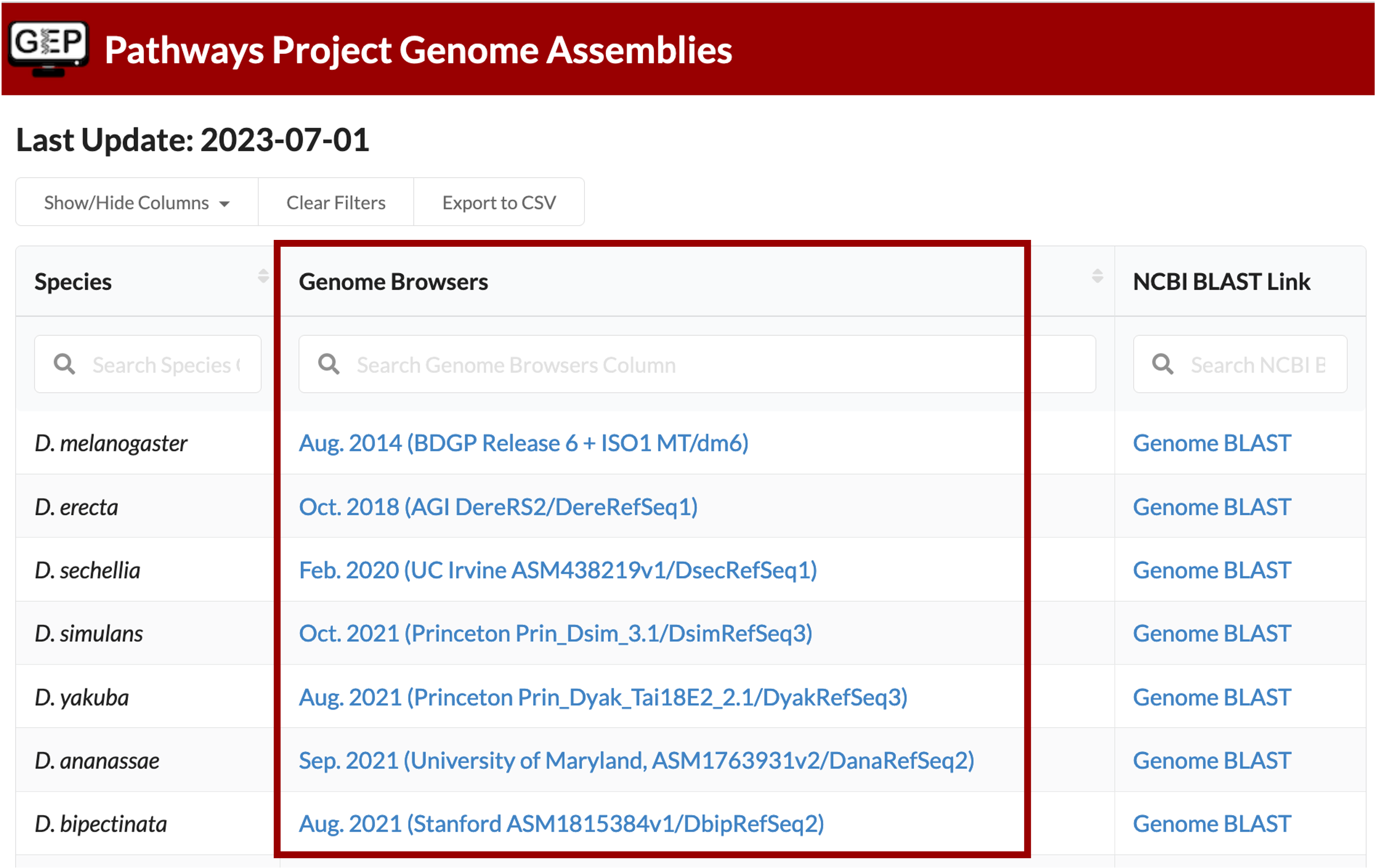
For example, D. yakuba has three assembly options to choose from and according to the Genome Assemblies page, we should use the “Aug. 2021 (Princeton Prin_Dyak_Tai18E2_2.1/ DyakRefSeq3)” assembly when annotating D. yakuba.