During Summer 2021, five Summer Fellows from New Jersey City University (NJCU), Rutgers University, and Washington University in St. Louis (WUSTL) reconciled coding region annotation projects from the Drosophila ananassae F element, the D. ananassae D element, and the D. bipectinata F element. The five Summer Fellows (Ishtar Olaveja, Jackie Hester, Annabelle Laughlin, Martin Dalling, and Alice Herrmann) were mentored by Dr. Cindy J. Arrigo at NJCU with support from Wilson Leung at WUSTL.
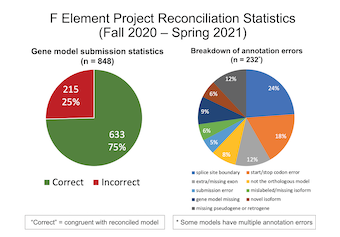
Collectively, the Summer Fellows reconciled 848 gene models and completed 21 unique TSS annotations. Approximately 75% of the submitted gene models were in congruence with the final gene models. The most common annotation error in the submitted gene models is the selection of incorrect splice site boundaries (24%). The image above illustrates the preliminary reconciliation statistics for the F element projects in Summer 2021. We anticipate that the final reconciliation statistics will be available in October 2021.
As part of reconciliation of the coding region annotation projects, the Summer Fellows also identified several interesting features on the F and D elements. These interesting features include a novel isoform for the Zyx gene and a partial duplication of Arl4 on the D. ananassae F element, three novel paralogs of a male-specific gene in D. ananassae (derived from CG3795), and a retrotransposed pseudogene derived from yin on the D. bipectinata F element. The presentations which summarize their work this Summer are publicly available through Box.
Three of the Summer Fellows plan to continue their reconciliation work and the data analysis of the reconciled gene models this Fall. Some of the Summer Fellows also plan to present their work at local conferences and undergraduate research symposia at their local institutions.
The Summer Fellows were supported by a grant from the National Science Foundation (Award #: 2114661).